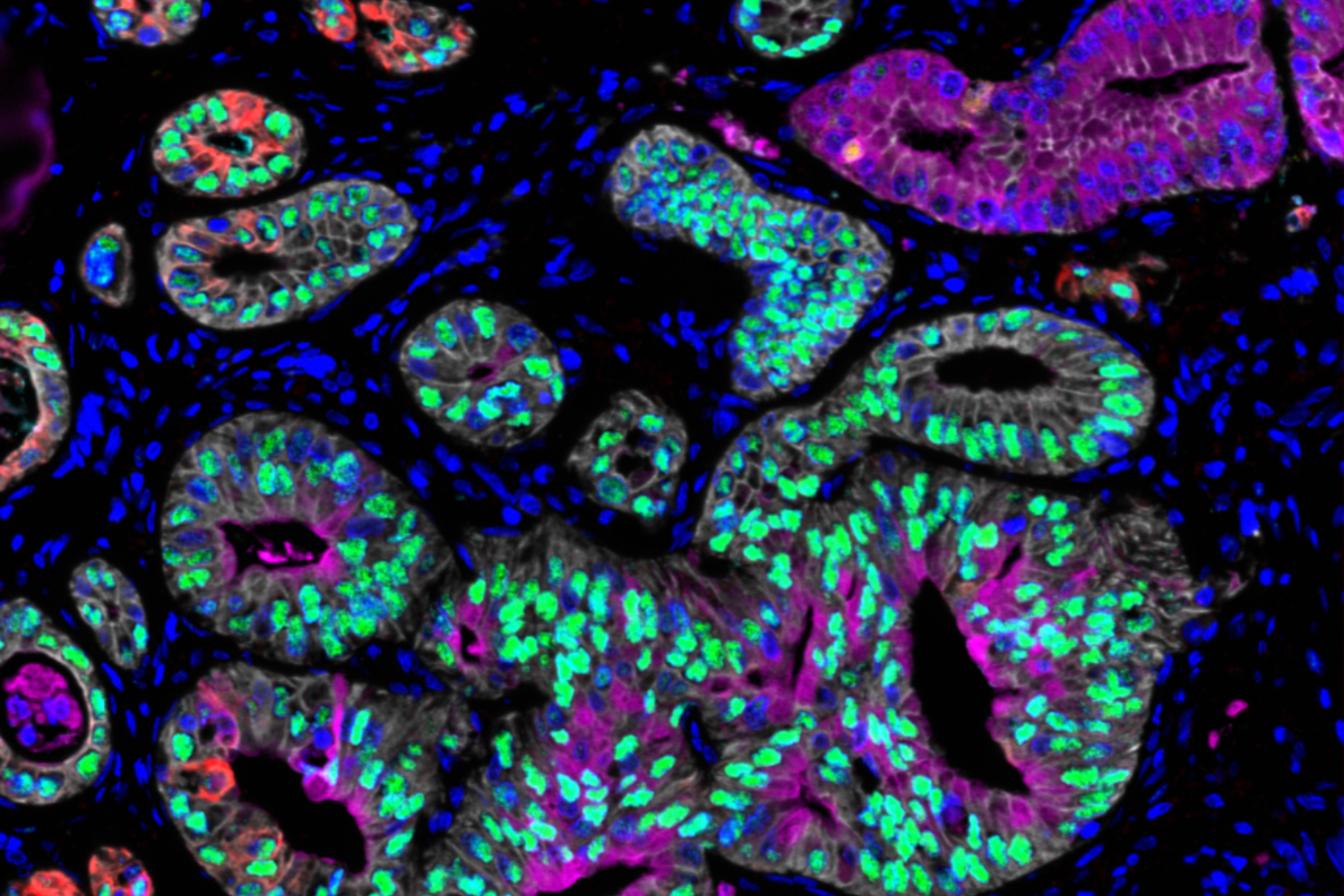
Measuring cancer cell state can reveal drug susceptibility
Study results also show that pancreatic tumor cells can be forced into a more susceptible state by changing their environment.
Over the past few decades, scientists have made great strides in understanding the genetic mutations that can drive cancer. For some types of cancer, these discoveries have led to the development of drugs that target specific mutations.
However, there are still many types of cancer for which no such targeted therapies are available. A team of researchers from MIT, Dana Farber Cancer Institute, and other institutions is now examining whether another cell trait — RNA expression patterns — influences drug responses and can be used to identify treatments a tumor might be susceptible to.
In a new study of pancreatic cancer cells, the researchers identified three prototypical RNA-expression states and uncovered differences in their susceptibility to a variety of cancer drugs. They also discovered that altering the tumor microenvironment can drive tumor cells from one state to another, potentially offering a way to make them more susceptible to a particular drug.
“What we show in this paper is that cancer cell state is plastic in response to the microenvironment and has a dramatic impact on drug sensitivity. This opens new frontiers for thinking about drug development and how to select drugs for individual patients,” says Alex Shalek, a core member of the Institute for Medical Engineering and Science (IMES) at MIT, an associate professor of chemistry, and an extramural member of MIT’s Koch Institute for Integrative Cancer Research. He is also a member of the Ragon Institute of MGH, MIT and Harvard and an institute member of the Broad Institute.
Shalek and Brian Wolpin, an associate professor of medicine at Harvard Medical School and Dana-Farber Cancer Institute; William Hahn, a professor of medicine at Harvard Medical School and Dana-Farber; and Andrew Aguirre, an assistant professor of medicine at Harvard Medical School and Dana-Farber; are the senior authors of the study, which appears today in Cell. The paper’s lead authors are Srivatsan Raghavan, an instructor in medicine at Harvard Medical School and Dana-Farber; Peter Winter, an MIT postdoc; Andrew Navia, an MIT graduate student; and Hannah Williams, a research fellow in medicine at Harvard Medical School and Dana-Farber.
Cell states
Sequencing a cell’s genome can reveal cancer-linked mutations, but identifying these mutations doesn’t always provide information that can be acted upon to treat a particular tumor. To generate additional data that could be used to help choose more targeted treatments, Shalek and other researchers have turned to single-cell RNA-sequencing, which reveals the genes that are being expressed by each cell at a moment in time.
“There are plenty of situations where the genetics are incredibly important, where you can develop these very precise drugs that target mutations or translocations,” Navia says. “But in many instances mutations alone don’t give you an effective way to target cancer cells relative to healthy ones.”
In this study, the researchers analyzed cells from pancreatic ductal adenocarcinoma (PDAC). There are very few targeted drugs available to treat pancreatic tumors, so most patients receive chemotherapy drugs that may be effective initially but often stop working as tumors become resistant. Using single-cell RNA-sequencing, the researchers analyzed about 25 metastatic tumor samples from pancreatic cancer patients.
Previous analyses of pancreatic tumor cell RNA have revealed two broad categories of cell states: basal-like, which is a more aggressive state, and classical. In the new study, the researchers identified a third state that appears to be an intermediate between those two. Cancer cells may pass through this state as they transition from classical to basal-like, the researchers say.
The researchers also found that the environment in which cancer cells are grown plays a key role in determining their state. In this study, they grew matched “organoids,” or tiny cancer aggregates from each patient’s biopsy. Such organoids are often used in precision medicine pipelines to model tumors from individual patients, to help identify drugs that might be useful for those individuals.
When comparing each in vivo single-cell profile to the matched ex vivo organoid model, the researchers found that the organoids often exist in a different RNA state than cancer cells examined directly from the same patient. “We see the same DNA mutations in the original tumor and its model, but when we start to examine what they look like at the RNA level, we find that they’re very, very different,” Shalek says.
That suggests that the state of a tumor can be influenced by the conditions in which it’s grown rather than its genetics alone, he says. The researchers also found that they could drive cancer cells, even long-established cell line models, to switch between different states by changing their growth conditions. Treating cells with TGF-beta, for example, drives them to a more aggressive, basal-like state, while taking TGF-beta away leads the cells to revert to the classical state in a dish.
Cells in each of those states depend on different cell-signaling pathways to survive, so knowing the cell state is critical to selecting the right kind of drug to treat a particular tumor, the researchers say.
“When we started looking at drug sensitivity, it became very clear that the same model pushed into a different state would respond very differently to a drug,” Navia says. “These state-specific sensitivities become critical as we think about selecting drugs and avoiding resistance. If you don’t know the right state, you could pick the entirely wrong compound and try to target the wrong pathways. If you don’t consider plasticity, the cancer may only respond temporarily until its cells change state.”
Targeted therapy
The findings suggest that further analyzing the interplay of genetics, cell state, and the tumor microenvironment could help researchers to develop new drugs that would effectively target individual patients’ tumors.
“We’re not erasing decades of understanding cancer as a genetic disease, but we’re certainly saying that we need to much better understand the intersection between genetics and state,” Winter says. “Cell state absolutely has ties to the underlying sensitivity of certain models, and therefore patients and to specific drugs.”
The discovery that cancer cells can be driven from one state to another by modifying the signals in their microenvironment raises the possibility of locking cancer cells into a particular state in a predictable way by therapeutically altering the tumor microenvironment, and then giving a separate drug to target that locked state and enhance treatment efficacy.
With their colleagues at Dana-Farber, the MIT team is now running much larger drug screens to measure how each drug affects pancreatic cancer cells in different states. They are also studying other types of cancer to determine if those cancer cells are also able to transition between different states in response to changes in their microenvironment.
The research was funded, in part, by the National Institutes of Health, the Koch Institute and Dana-Farber/Harvard Cancer Center Bridge Project, the Ludwig Center for Molecular Oncology at MIT, the Beckman Young Investigator Program, a Sloan Fellowship in Chemistry, and the Pew-Stewart Scholars Program for Cancer Research.