Shalek paper published in Nature Communications
The paper, Live cell tagging tracking and isolation for spatial transcriptomics using photoactivatable cell dyes, was published online on August 17, 2021.
A paper authored by Alex S Genshaft, Carly G. K. Ziegler, Constantine N. Tzouanas, Benjamin E. Mead, Alex M. Jaeger, Andrew W. Navia, Ryan P. King, Miyeko D. Mana, Siyi Huang, Vanessa Mitsialis, Scott B. Snapper, Ömer H. Yilmaz, Tyler Jacks, Jeffrey F. Van Humbeck and Alex K. Shalek was published in Nature Communications on August 17, 2021.
Live cell tagging tracking and isolation for spatial transcriptomics using photoactivatable cell dyes
Alex S Genshaft, Carly G. K. Ziegler, Constantine N. Tzouanas, Benjamin E. Mead, Alex M. Jaeger, Andrew W. Navia, Ryan P. King, Miyeko D. Mana, Siyi Huang, Vanessa Mitsialis, Scott B. Snapper, Ömer H. Yilmaz, Tyler Jacks, Jeffrey F. Van Humbeck and Alex K. Shalek
Department of Chemistry, Massachusetts Institute of Technology, Cambridge MA, USA.
Nat Commun 12, 4995 (2021)
DOI: 10.1126/sciadv.aaz3168
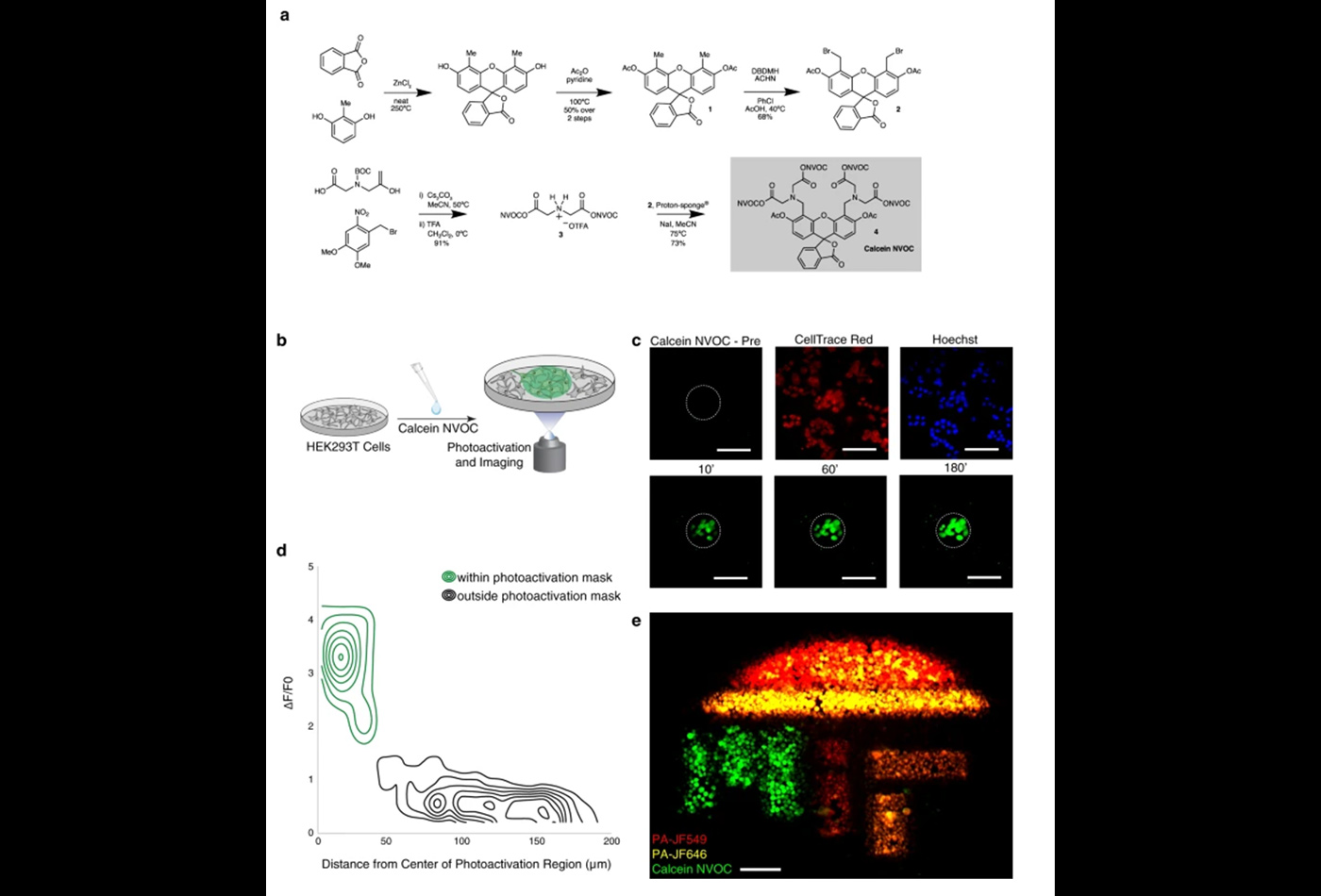
A cell’s phenotype and function are influenced by dynamic interactions with its microenvironment. To examine cellular spatiotemporal activity, we developed SPACECAT—Spatially PhotoActivatable Color Encoded Cell Address Tags—to annotate, track, and isolate cells while preserving viability. In SPACECAT, samples are stained with photocaged fluorescent molecules, and cells are labeled by uncaging those molecules with user-patterned near-UV light. SPACECAT offers single-cell precision and temporal stability across diverse cell and tissue types. Illustratively, we target crypt-like regions in patient-derived intestinal organoids to enrich for stem-like and actively mitotic cells, matching literature expectations. Moreover, we apply SPACECAT to ex vivo tissue sections from four healthy organs and an autochthonous lung tumor model. Lastly, we provide a computational framework to identify spatially-biased transcriptome patterns and enriched phenotypes. This minimally perturbative and broadly applicable method links cellular spatiotemporal and/or behavioral phenotypes with diverse downstream assays, enabling insights into the connections between tissue microenvironments and (dys)function.
Read the Full Text at Nature Communications.
The Shalek Lab creates and implements new approaches to elucidate cellular and molecular features that inform tissue-level function and dysfunction across the spectrum of human health and disease.