Newly discovered enzyme “square dance” helps generate DNA building blocks
MIT biochemists can trap and visualize an enzyme as it becomes active — an important development that may aid in future drug development.
How do you capture a cellular process that transpires in the blink of an eye? Biochemists at MIT have devised a way to trap and visualize a vital enzyme at the moment it becomes active — informing drug development and revealing how biological systems store and transfer energy.
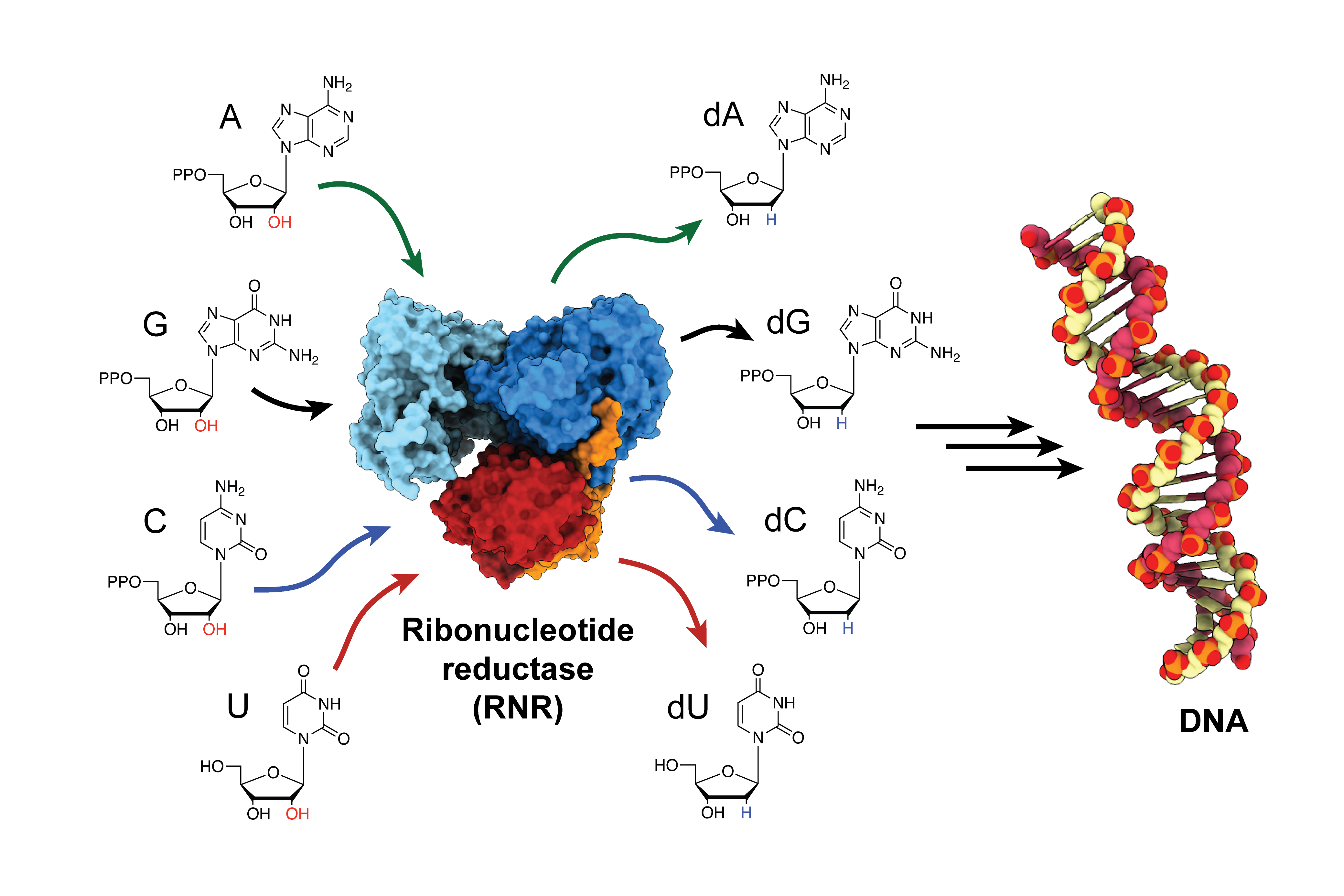
The enzyme, ribonucleotide reductase (RNR), is responsible for converting RNA building blocks into DNA building blocks, in order to build new DNA strands and repair old ones. RNR is a target for anti-cancer therapies, as well as drugs that treat viral diseases like HIV/AIDS. But for decades, scientists struggled to determine how the enzyme is activated because it happens so quickly. Now, for the first time, researchers have trapped the enzyme in its active state and observed how the enzyme changes shape, bringing its two subunits closer together and transferring the energy needed to produce the building blocks for DNA assembly.
Before this study, many believed RNR’s two subunits came together and fit with perfect symmetry, like a key into a lock. “For 30 years, that’s what we thought,” says Catherine Drennan, an MIT professor of chemistry and biology and a Howard Hughes Medical Institute investigator. “But now, we can see the movement is much more elegant. The enzyme is actually performing a ‘molecular square dance,’ where different parts of the protein hook onto and swing around other parts. It’s really quite beautiful.”
Drennan and JoAnne Stubbe, professor emerita of chemistry and biology at MIT, are the senior authors on the study, which appeared in the journal Science on March 26. Former graduate student Gyunghoon “Kenny” Kang PhD ’19 is the lead author.
All proteins, including RNR, are composed of fundamental units known as amino acids. For over a decade, Stubbe’s lab has been experimenting with substituting RNR’s natural amino acids for synthetic ones. In doing so, the lab realized they could trap the enzyme in its active state and slow down its return to normal. However, it wasn’t until the Drennan lab gained access to a key technological advancement — cryo-electron microscopy — that they could snap high-resolution images of these “trapped” enzymes from the Stubbe lab and get a closer look.
“We really hadn’t done any cryo-electron microscopy at the point that we actively started trying to do the impossible: get the structure of RNR in its active state,” Drennan says. “I can’t believe it worked; I’m still pinching myself.”
The combination of these techniques allowed the team to visualize the complex molecular dance that allows the enzyme to transport the catalytic “firepower” from one subunit to the next, in order to generate DNA building blocks. This firepower is derived from a highly reactive unpaired electron (a radical), which must be carefully controlled to prevent damage to the enzyme.
According to Drennan, the team “wanted to see how RNR does the equivalent of playing with fire without getting burned.”
First author Kang says slowing down the radical transfer allowed them to observe parts of the enzyme no one had been able to see before in full. “Before this study, we knew this molecular dance was happening, but we’d never seen the dance in action,” he says. “But now that we have a structure for RNR in its active state, we have a much better idea about how the different components of the enzyme are moving and interacting in order to transfer the radical across long distances.”
Although this molecular dance brings the subunits together, there is still considerable distance between them: The radical must travel 35-40 angstroms from the first subunit to the second. This journey is roughly 10 times farther than the average radical transfer, according to Drennan. The radical must then travel back to its starting place and be stored safely, all within a fraction of a second before the enzyme returns to its normal conformation.
Because RNR is a target for drugs treating cancer and certain viruses, knowing its active-state structure could help researchers devise more effective treatments. Understanding the enzyme’s active state could also provide insight into biological electron transport for applications like biofuels. Drennan and Kang hope their study will encourage others to capture fleeting cellular events that have been difficult to observe in the past.
“We may need to reassess decades of past results,” Drennan says. “This study could open more questions than it answers; it’s more of a beginning than an end.”
This research was funded by the National Institutes of Health, a David H. Koch Graduate Fellowship, and the Howard Hughes Medical Institute.