Combating COVID-19 with Chemistry
Despite MIT's desolate campus, members of the Hong, Kiessling, Pentelute, Shalek, and Swager labs continue to pursue a solution to the global pandemic.
In March of this year, MIT’s open Cambridge campus, typically abuzz with tourists and visitors in addition to members of the institute’s community, became a ghost town practically overnight. In a matter of days, students were sent home, classes were moved online, and the community shifted to an entirely work-from-home format for the vast majority of staff, students, faculty, and researchers in response to the COVID-19 global pandemic. As most of the world stayed inside, sheltering in place, venturing out of their self-isolation only for essential food and supply runs, a select few members of the Department of Chemistry – those in a race to discover possible solutions to the world’s viral crisis – were granted special permission to carry on their research on campus at a responsible social distance. Efforts from the labs of Professor Mei Hong, Professor Laura L. Kiessling, Professor Bradley L. Pentelute, Professor Alex K. Shalek, and Professor Timothy Manning Swager aim to contribute to resolving COVID-19.
The Hong Group

Since mid-March, Professor Mei Hong’s research group has directed its membrane protein expertise to investigate the structure of an essential SARS-COV-2 protein, the envelope protein E. The E protein is one of the three SARS coronavirus membrane proteins, and is involved in several aspects of the coronavirus lifecycle, including virus assembly, budding, and pathogenesis. The protein forms a cation channel in the lipid membrane, which can be blocked by several small-molecule drugs. In addition it causes membrane curvature, which is required for releasing new progeny viruses from the Golgi membrane of the cell. The envelope protein’s channel activity has been implicated in the ability of SARS-COV-2 to over-stimulate the inflammation pathways of the host cell, with deadly consequences. Deletion of the E gene from the virus has been shown to reduce virus titers, attenuate infection in animal models, and confer protection in hamsters and mice.
Currently, little is known about the three-dimensional structure of the E protein in lipid membranes. Knowing this structure will allow rational design of E-targeting antiviral drugs to stop virus propagation. The Hong group is cloning and purifying the E protein and will employ multidimensional NMR spectroscopy to determine its membrane-bound structure.
The Kiessling Group

A feature of enveloped viruses, including coronaviruses, is their high levels of glycosylation. Viral glycosylation plays multiple roles in virus pathology, and infection by the novel SARS-CoV-2, the virus that causes COVID-19, is mediated by the viral spike glycoprotein (S). Viruses have glycosylation patterns that differ from those of the host, so they can be recognized by host proteins. Professor Laura Kiessling and her research group are investigating whether human carbohydrate-binding proteins (lectins) limit viral infection. Human lectins are present at mucosal barriers, including in the lung. The levels of these proteins can differ between individuals in response to age or polymorphisms (changes in the genetic coding) that alter their production or stability.
The Kiessling Group anticipates that these lectins can function in vitro as tools to capture virus and in vivo as immunomodulators. Thus, understanding those lectins that can bind coronaviruses from human patients will lay the groundwork to elucidate fundamental aspects of human immune responses to the virus. Moreover, because the lectins the group proposes to examine are human proteins, they do not expect them to have problematic effects. If they block viral infectivity (or effectively modulate immune responses), they are unlikely to have major deleterious effects such as toxicity or immunogenicity.
The group has made some models that suggest the lectins have the right alignment:
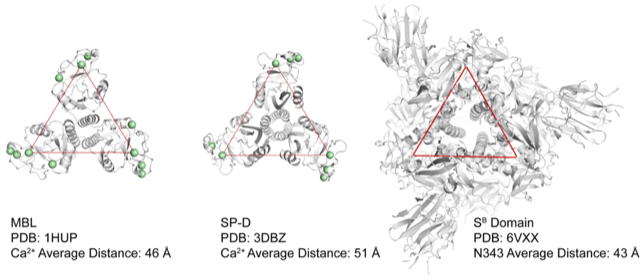
Spike protein showing the distance between glycosylation sites (the protein is a trimer on the right) compared to two human lectins (SP-D and MBL) with the location of their carbohydrate binding sites highlighted. This computational model is tantalizing in that the spacing looks excellent for binding. These images were made by postdoctoral researcher Dr. Mike Wuo, who is involved with the project.
The Pentelute Group

Researchers in Professor Brad Pentelute’s lab are focused on delivering safe and effective peptides for prophylactic treatment and rapid early therapeutic intervention against COVID-19 infection. They approach this goal by design of peptide binders specific to the spike protein of the SARS-CoV-2 coronavirus. Disrupting the SARS-CoV-2 spike protein interaction with its human cell membrane receptor can prevent virus cell entry, offering a promising therapeutic modality.
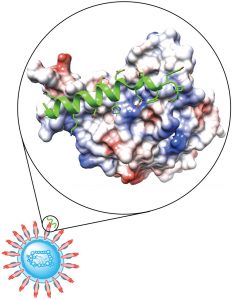
With collaborators in the tenOever lab at Icahn School of Medicine at Mount Sinai and the Hacohen lab at the Broad Institute, they are testing the activities of these peptide binders in live- and pseudo-virus inhibition assays, respectively. A path to clinical trials was identified in collaboration with Dr. Robert Shoemaker at the National Institute of Health, in which the most potent and least toxic peptide will be produced under GMP conditions and formulated in nasal and spray inhalers for evaluation in humans. The Pentelute Lab approach will ensure COVID-19 treatment by blocking SARS-CoV-2 interaction with human cells.
The Shalek Group

Professor Alex Shalek’s group has three projects pertaining to COVID-19. First, they are studying the cellular correlates and pathophysiology of novel therapeutics and vaccines against SARS-CoV-2. Next, they are working to identify an actionable host-pathogen interplay, and finally, they hope to understand the correlates of a clinical outcome.
The lab seeks to fundamentally transform the study and engineering of human immune responses. To do this, they create and implement cutting-edge experimental and computational platforms to elucidate cellular and molecular features that inform tissue-level (dys)function across the spectrum of human health and disease. The team is actively working on three major COVID-19 related projects at MIT, the Ragon Institute, and the Broad Institute, and helping with others. First, with partners at the Wyss Institute, Brigham and Women’s Hospital, and the Icahn School of Medicine at Mount Sinai, they are examining whether different experimental antiviral treatments and current FDA approved drugs can be used to make host cells less amenable to viral entry and release for rapid repurposing. Second, with members of the Human Cell Atlas Lung Biological Network, they are examining which specific cells in different tissues become infected with SARS-CoV-2 to uncover targetable host factors for controlling infection. And, third, the team is working with partners around Boston and the world to understand why some individuals with pre-existing conditions (e.g. high blood pressure, cancer) or risk factors develop severe disease while some populations (e.g. pediatric) are seemingly resistant to severe infection to realize better treatments and cures.
The Swager Group

Professor Tim Swager’s lab has developed new biosensors that make use of dynamic droplets dispersed in water. These droplets have multiple different internal structures that allow them to behave as micro-lens systems directing light either transmitted through them or light coming from fluorescent dyes within the droplets. Placement of antibodies and recognition elements at the surface of the droplets and when they interact with viruses, proteins, or bacteria, the droplets change their organization in a way that creates a large scattering of light or directional light emission. These signals allow for a very strong signal to be generated by binding to a single virus and detection can be performed with common smartphones. Using the recognition elements developed by Professor Hadley Sikes of MIT’s Mechanical Engineering department, Swager and his coworkers are developing an ultra-sensitive rapid sensor for COVID-19 and the antibodies to this virus.
As humanity continues to await the end of the pandemic and the beginning of living in coexistence with this virus, the Department of Chemistry is proud to be home to those who are contributing to a solution to this crisis.