Griffin paper published in Science Advances
The paper, Three-spin solid effect and the spin diffusion barrier in amorphous solids, was published online on July 26, 2019.
A paper authored by Kong Ooi Tan, Michael Mardini, Chen Yang, Jan Henrik Ardenkjær-Larsen, and Professor Robert Guy Griffin was published in Science Advances on July 26, 2019.
Three-spin solid effect and the spin diffusion barrier in amorphous solids
Kong Ooi Tan, Michael Mardini, Chen Yang, Jan Henrik Ardenkjær-Larsen, and Robert G. Griffin
Science Advances, July 26, 2019
Vol. 5, no. 7, eaax2743
DOI: 10.1126/sciadv.aax2743
Dynamic nuclear polarization (DNP) is a technique that enables NMR signal intensities to be boosted by a factor of 100 or more, leading to a time savings of ≳10,000. This is important because it allows an NMR experiment that would otherwise take approximately 30 years to complete to be performed in a day.
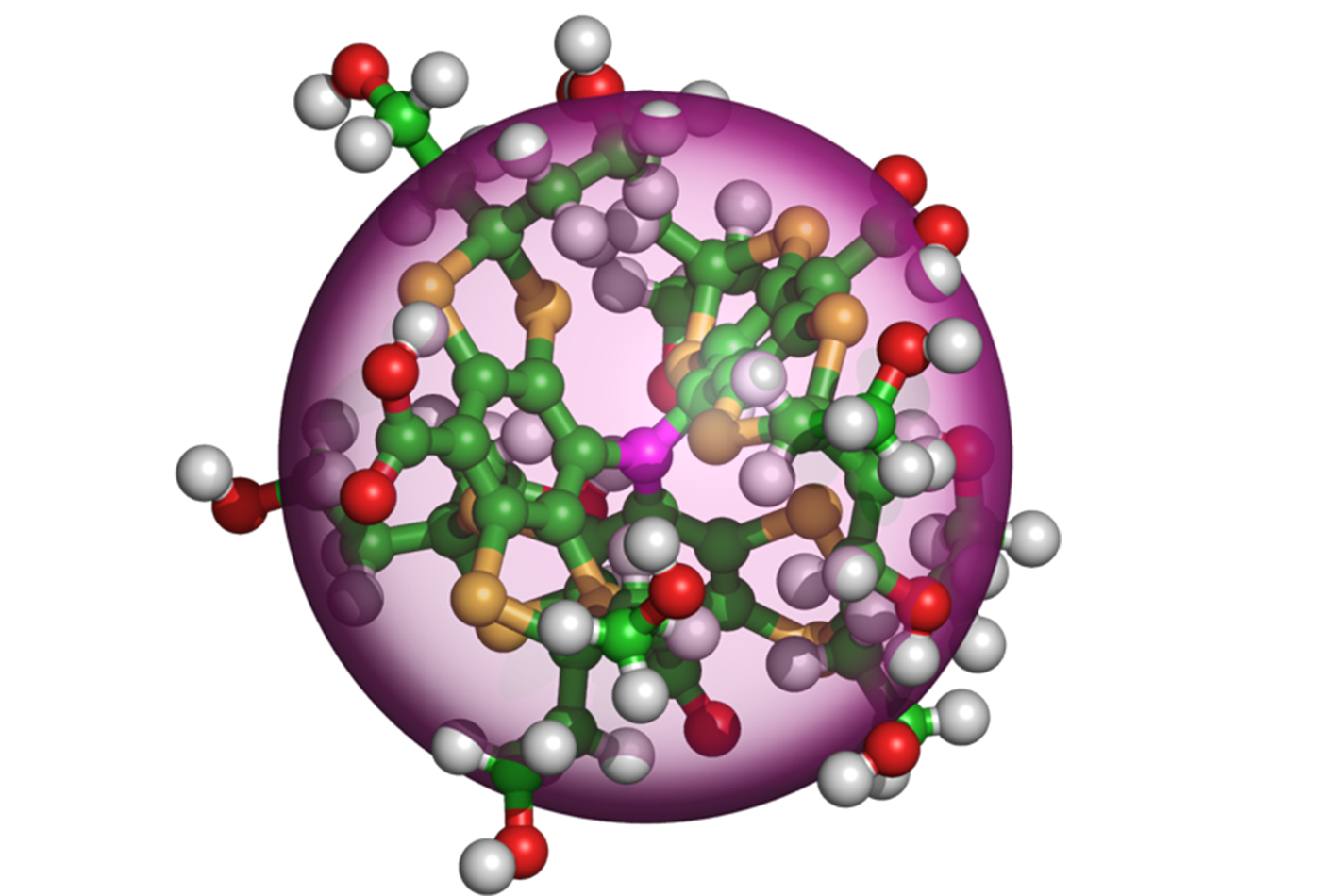
The signal enhancement is accomplished by transferring polarization from a paramagnetic molecule, a polarizing agent, to the NMR-active nuclei of nearby molecules. The nearby polarized molecules then disperse the polarization to more distant molecules via a mechanism known as nuclear spin diffusion. However, the conventional wisdom states that not all nearby molecules participate in the relay process because their proximity to the polarizing agent results in an interaction that quenches spin diffusion. These molecules are said to reside inside the spin diffusion barrier (SDB). Recent theoretical calculations suggest that the SDB can have a radius of 20-40 Å; however, experimental data on the size of this barrier is very limited.
In this study in Science Advances, researchers exploit the three-spin solid effect (TSSE) to determine the size of the SDB. They show that for the trityl molecule, a common organic polarizing agent, the SDB resides at the edge of the molecular structure (<6 Å). Therefore, all nearby molecules participate in spin diffusion. This first and unexpected experimental result for the case of trityl shows there is not a SDB, and provides insight into the details of the DNP process. Furthermore, the results will inform more accurate modeling of the spin diffusion processes that are used to extract structural information from pharmaceutical products, new materials, and biological systems such as membrane proteins and amyloid fibrils.
Read the Full Text at Science Advances.
A large fraction of the Griffin Group‘s research effort is devoted to the development of new magnetic resonance techniques to study molecular structure and dynamics