New Computational Method from the Wang Lab Published in Nature Methods
The paper, “Search and match across spatial omics samples at single-cell resolution,” was published on September 18, 2024.
Spatial Transcriptomics (ST) was crowned as the Technology of the Year by Nature Methods in 2020. This cutting-edge technology allows for the detection of gene expression within cells while preserving their “spatial information”—meaning the exact physical location of each cell within the tissue. This spatial information is essential for understanding how cells interact within their native environments, offering insights that traditional gene expression technologies, such as single-cell RNA-seq, which may lose positional context, cannot provide. Up to now, various experimental technologies have been developed based on different mechanisms, to expand the ability to measure molecular characteristics beyond gene expression, such as RNA translation expression and protein abundance. However, the integration and comparison of spatial data across different technologies and molecular features pose significant challenges due to variations in technologies, sample types, resolutions, and gene sets.
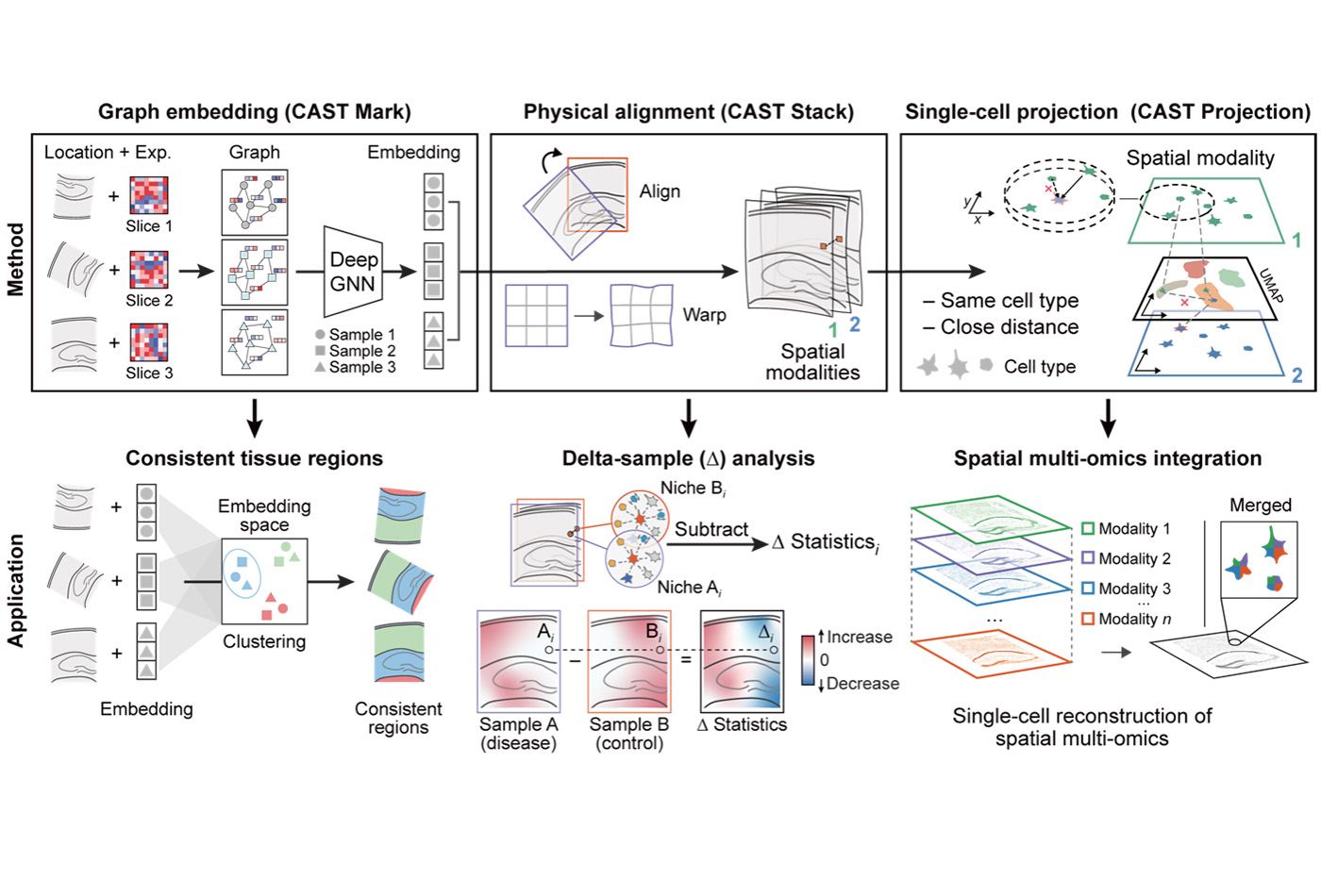
To address these challenges, researchers from Associate Professor of Chemistry Xiao Wang’s lab at MIT and the Broad Institute developed a computational method called CAST. CAST uses deep graph neural networks—advanced machine learning algorithms designed to model relationships between data points, in this case, individual cells, based on their connections (or “edges”) within a spatial network. CAST can extract shared spatial features, enabling spatial-to-spatial searches and physical alignment at the single-cell level.
Since CAST can physically align samples, enabling them to be mapped into a unified spatial framework—referred to as the ‘consistent spatial coordinate system’— the researchers used this system as “Google image search” to align new query data against reference spatial atlases. They also applied CAST to identify spatial pathological patterns in Alzheimer’s disease. They compared the gene expression and cell-to-cell interactions in diseased and healthy brain tissue slices at a high spatial resolution. Similarly, they also uncovered unique differential patterns during the tissue regeneration process in the injured axolotl brain by aligning samples from injured and healthy brain hemispheres. Moreover, CAST was employed to reconstruct cells with independent measurements of spatially-resolved transcriptomes (RNA expression data) and translatomes (RNA translation data). This allowed them to generate single-cell relative translation efficiency (scRTE) profiles—measurements of how efficiently different cell types translate RNA into proteins across various brain regions—thereby revealing variations of translation efficiency across different cell types and brain regions.
As an integrative framework, CAST facilitates seamless single-cell spatial data comparisons across technologies, modalities, and sample conditions. This work undoubtedly enhances the strategies for analyzing spatial transcriptome data, unlocking the potential of this rapidly advancing technology and the growing volume of spatial data. Moreover, the proposed strategy provides valuable guidance for researchers in designing experiments, particularly in sample collection. CAST might play a significant role in the ongoing development of spatial transcriptomics.
The Wang Group is developing and applying the state-of-the-art tools of chemistry, biophysics and genomics to map and perturb biological tissues from molecules to systems. In specific, with in situ sequencing of nucleic acids as the core approaches, the group aims to develop high-resolution and highly-multiplexed molecular imaging methods across multiple scales toward understanding the physical and chemical basis of brain wiring and function.